| Version 9 (modified by , 8 years ago) ( diff ) |
|---|
Random Forest
I am using Ranger a fast implementation of the random forest machine learning algorithm. The source code can be grabbed from here: https://github.com/imbs-hl/ranger or cloned from GitHub via
git clone https://github.com/imbs-hl/ranger.git
To compile Range then do
cd ranger/cpp_version mkdir build cd build cmake .. make
Here is the full output (on an Ubuntu 18.04)
user@machine:~> git clone https://github.com/imbs-hl/ranger.git Cloning into 'ranger'... remote: Counting objects: 8000, done. remote: Compressing objects: 100% (32/32), done. remote: Total 8000 (delta 6), reused 5 (delta 3), pack-reused 7965 Receiving objects: 100% (8000/8000), 19.88 MiB | 2.30 MiB/s, done. Resolving deltas: 100% (6172/6172), done. user@machine:~> cd ranger user@machine:~/ranger> cd cpp_version/ user@machine:~/ranger/cpp_version> mkdir build user@machine:~/ranger/cpp_version> cd build user@machine:~/ranger/cpp_version/build> cmake .. -- The C compiler identification is GNU 7.3.0 -- The CXX compiler identification is GNU 7.3.0 -- Check for working C compiler: /usr/bin/cc -- Check for working C compiler: /usr/bin/cc -- works -- Detecting C compiler ABI info -- Detecting C compiler ABI info - done -- Detecting C compile features -- Detecting C compile features - done -- Check for working CXX compiler: /usr/bin/c++ -- Check for working CXX compiler: /usr/bin/c++ -- works -- Detecting CXX compiler ABI info -- Detecting CXX compiler ABI info - done -- Detecting CXX compile features -- Detecting CXX compile features - done CMake Warning (dev) at CMakeLists.txt:2 (cmake_minimum_required): Compatibility with CMake < 2.4 is not supported by CMake >= 3.0. This warning is for project developers. Use -Wno-dev to suppress it. -- Performing Test COMPILER_SUPPORTS_CXX11 -- Performing Test COMPILER_SUPPORTS_CXX11 - Success Compiler with C++11 support found. -- Configuring done -- Generating done -- Build files have been written to: /home/fact/ranger/cpp_version/build user@machine:~/ranger/cpp_version/build> make -j8 Scanning dependencies of target ranger [ 23%] Building CXX object CMakeFiles/ranger.dir/src/Forest/ForestProbability.o [ 23%] Building CXX object CMakeFiles/ranger.dir/src/Forest/ForestClassification.o [ 29%] Building CXX object CMakeFiles/ranger.dir/src/Forest/ForestSurvival.o [ 23%] Building CXX object CMakeFiles/ranger.dir/src/Forest/Forest.o [ 23%] Building CXX object CMakeFiles/ranger.dir/src/Forest/ForestRegression.o [ 35%] Building CXX object CMakeFiles/ranger.dir/src/Tree/Tree.o [ 41%] Building CXX object CMakeFiles/ranger.dir/src/Tree/TreeClassification.o [ 47%] Building CXX object CMakeFiles/ranger.dir/src/Tree/TreeProbability.o [ 52%] Building CXX object CMakeFiles/ranger.dir/src/Tree/TreeRegression.o [ 58%] Building CXX object CMakeFiles/ranger.dir/src/Tree/TreeSurvival.o [ 64%] Building CXX object CMakeFiles/ranger.dir/src/main.o [ 70%] Building CXX object CMakeFiles/ranger.dir/src/utility/ArgumentHandler.o [ 76%] Building CXX object CMakeFiles/ranger.dir/src/utility/Data.o [ 82%] Building CXX object CMakeFiles/ranger.dir/src/utility/DataChar.o [ 88%] Building CXX object CMakeFiles/ranger.dir/src/utility/DataFloat.o [ 94%] Building CXX object CMakeFiles/ranger.dir/src/utility/utility.o [100%] Linking CXX executable ranger [100%] Built target ranger user@machine:~/ranger/cpp_version/build>
Writing Input Files
Now we have the random forest, now we need the data. I assume that you have created a root-file with the simulation data you want to use already which is named simulation.root.
#include <iostream> #include <iomanip> #include <fstream> #include <TMath.h> #include <TChain.h> using namespace std; void writesim() { // Create chain for the tree Result // This is just easier than using TFile/TTree TChain c("Result"); // Add the input file to the c.AddFile("simulation.root"); // Define variables for all leaves to be accessed // By definition rootifysql writes only doubles double X, Y, MeanX, MeanY, Width, Length, CosDelta, SinDelta, M3Long, SlopeLong, Leakage1, SlopeSpreadWeighted, Size, ConcCore, ConcCOG, NumIslands, NumUsedPixels, Zd, Energy; // Connect the variables to the cordesponding leaves c.SetBranchAddress("X", &X); c.SetBranchAddress("Y", &Y); c.SetBranchAddress("MeanX", &MeanX); c.SetBranchAddress("MeanY", &MeanY); c.SetBranchAddress("Width", &Width); c.SetBranchAddress("Length", &Length); c.SetBranchAddress("CosDelta", &CosDelta); c.SetBranchAddress("SinDelta", &SinDelta); c.SetBranchAddress("M3Long", &M3Long); c.SetBranchAddress("SlopeLong", &SlopeLong); c.SetBranchAddress("Leakage1", &Leakage1); c.SetBranchAddress("NumIslands", &NumIslands); c.SetBranchAddress("NumUsedPixels", &NumUsedPixels); c.SetBranchAddress("Size", &Size); c.SetBranchAddress("Zd", &Zd); c.SetBranchAddress("Energy", &Energy); // Set some constants (they could be included in the database // in the future) double mm2deg = +0.0117193246260285378; // -------------------- Source dependent parameter calculation ------------------- ofstream fout0("sim-train.csv"); // %1 ofstream fout1("sim-test.csv"); // %0 ofstream fout2("sim-test-cuts.csv"); fout0 << "Energy Size Zd Dist Disp Slope M3L Leakage Width Length" << endl; fout1 << "Energy Size Zd Dist Disp Slope M3L Leakage Width Length" << endl; fout2 << "Energy Size Zd Dist Disp Slope M3L Leakage Width Length" << endl; // Loop over all wobble positions in the camera for (int i=0; i<c.GetEntries(); i++) { // read the i-th event from the file c.GetEntry(i); // First calculate all cuts to speedup the analysis double area = TMath::Pi()*Width*Length; int angle = 0; // -------------------- Source dependent parameter calculation ------------------- double cr = cos(angle*TMath::DegToRad()); double sr = sin(angle*TMath::DegToRad()); double px = cr*X-sr*Y; double py = cr*Y+sr*X; double dx = MeanX - px*1.02; double dy = MeanY - py*1.02; double norm = sqrt(dx*dx + dy*dy); double dist = norm*mm2deg; double lx = min(max((CosDelta*dy - SinDelta*dx)/norm, -1.), 1.); double ly = min(max((CosDelta*dx + SinDelta*dy)/norm, -1.), 1.); double alpha = asin(lx); double sgn = TMath::Sign(1., ly); // ------------------------------- Application ---------------------------------- double m3l = M3Long*sgn*mm2deg; double slope = SlopeLong*sgn/mm2deg; // --------------------------------- Analysis ----------------------------------- double xi = 1.340 + 0.0755*slope + 1.67972*(1-1/(1+4.86232*Leakage1)); double sign1 = m3l+0.07; double sign2 = (dist-0.5)*7.2-slope; double disp = (sign1<0 || sign2<0 ? -xi : xi)*(1-Width/Length); double thetasq = disp*disp + dist*dist - 2*disp*dist*sqrt(1-lx*lx); if (i%2==0) { fout0 << log10(Energy) << " "; fout0 << log10(Size) << " "; fout0 << Zd << " "; fout0 << dist << " "; fout0 << disp << " "; fout0 << slope << " "; fout0 << m3l << " "; fout0 << Leakage1 << " "; fout0 << Width << " "; fout0 << Length << endl; } else { fout1 << log10(Energy) << " "; fout1 << log10(Size) << " "; fout1 << Zd << " "; fout1 << dist << " "; fout1 << disp << " "; fout1 << slope << " "; fout1 << m3l << " "; fout1 << Leakage1 << " "; fout1 << Width << " "; fout1 << Length << endl; if (thetasq<0.024) continue; bool cutq = NumIslands<3.5 && NumUsedPixels>5.5 && Leakage1<0.1; if (!cutq) continue; bool cut0 = area < log10(Size)*898-1535; if (!cut0) continue; fout2 << log10(Energy) << " "; fout2 << log10(Size) << " "; fout2 << Zd << " "; fout2 << dist << " "; fout2 << disp << " "; fout2 << slope << " "; fout2 << m3l << " "; fout2 << Leakage1 << " "; fout2 << Width << " "; fout2 << Length << endl; } } }
Training
Now we train the random forest with the train data using ranger:
fact@ihp-pc45:~/Analysis> nice -n 10 ~/ranger-master/cpp_version/build/ranger --file sim-train.csv --depvarname Energy --memmode 1 --treetype 3 --verbose --impmeasure 1 --outprefix sim-train Starting Ranger. Loading input file: sim-train.csv. Growing trees .. Computing prediction error .. Tree type: Regression Dependent variable name: Energy Dependent variable ID: 0 Number of trees: 500 Sample size: 55417 Number of independent variables: 9 Mtry: 3 Target node size: 5 Variable importance mode: 1 Memory mode: 1 Seed: 0 Number of threads: 8 Overall OOB prediction error: 0.0178514 Saved variable importance to file sim-train.importance. Saved prediction error to file sim-train.confusion. Finished Ranger.
It will write a file called sim-train.forest with the forest and give you a list of the importance of the parameters
user@machine> cat ranger_out.importance Size: 2764.52 Zd: 1198.02 Dist: 695.544 Disp: 163.821 Slope: 246.754 M3L: 277.035 Leakage: 118.393 Width: 420.597 Length: 565.094
Testing
Now we can apply the forest to out test data. Once to all data and once to the data after background suppression:
nice -n 10 ~/ranger-master/cpp_version/build/ranger --file sim-test.csv --depvarname Energy --memmode 1 --treetype 3 --verbose --impmeasure 1 --predict sim-train.forest nice -n 10 ~/ranger-master/cpp_version/build/ranger --file sim-test-cuts.csv --depvarname Energy --memmode 1 --treetype 3 --verbose --impmeasure 1 --predict sim-train.forest
Here is an example output
Starting Ranger. Loading input file: sim-test-cuts.csv. Loading forest from file sim-train.forest. Predicting .. Tree type: Regression Dependent variable name: Energy Dependent variable ID: 0 Number of trees: 500 Sample size: 5135 Number of independent variables: 9 Mtry: 3 Target node size: 5 Variable importance mode: 1 Memory mode: 1 Seed: 0 Number of threads: 8 Saved predictions to file ranger_out.prediction. Finished Ranger.
Plotting Result
The predicted values have been written to ranger_out.prediction. We can read them now together with the test-data and do some control plots. Here is an example ROOT macro which plots the result for the test-data after background suppresion cuts.
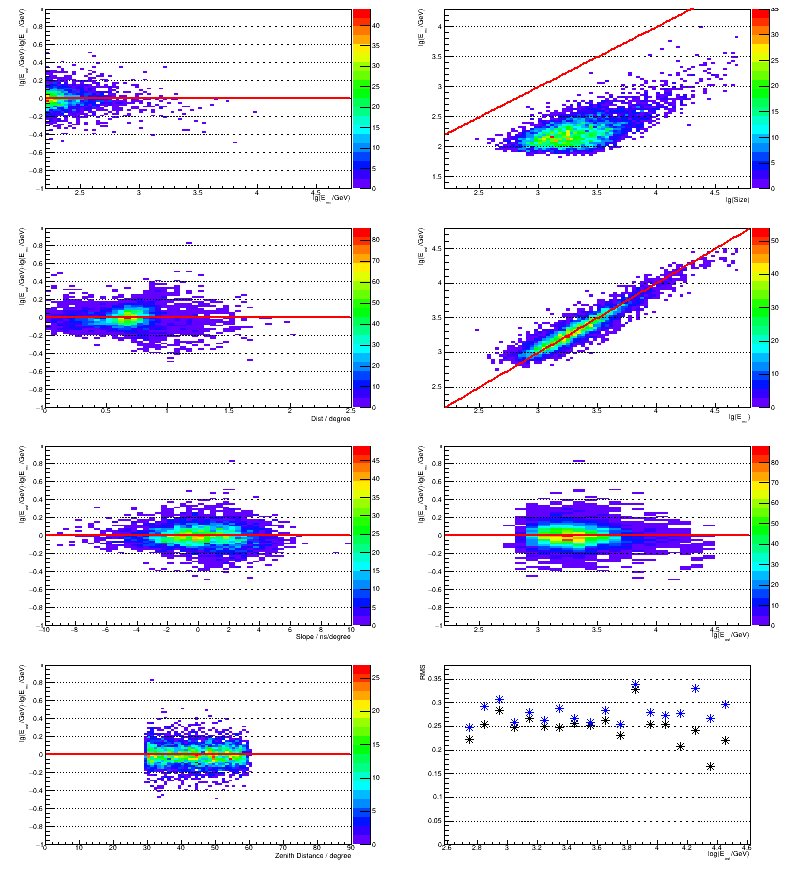
#include <fstream> #include <TH2.h> #include <TF1.h> #include <TGraph.h> #include <TCanvas.h> void readsim() { TH2F h2svse ("H_SvsE", "", 26*3, 2.2, 4.8, 3*30, 1.3, 4.3); TH2F h2est ("H_EstVsMC", "", 26*3, 2.2, 4.8, 26*3, 2.2, 4.8); TH2F h2bias ("H_BiasLog", "", 26*3, 2.2, 4.8, 100, -1, 1); TH2F h2lin ("H_BiasLin", "", 26*3, 2.2, 4.8, 100, -1, 2); TH2F h2size ("H_ResSize", "", 26*3, 2.2, 4.8, 100, -1, 1); TH2F h2dist ("H_ResDist", "", 50, 0, 2.5, 100, -1, 1); TH2F h2slope("H_ResSlope", "", 50, -10, 10, 100, -1, 1); TH2F h2zd ("H_ResZd", "", 90, 0, 90, 100, -1, 1); ifstream fin0("sim-test-cuts.csv"); ifstream fin1("ranger_out.prediction"); TString str; str.ReadLine(fin0); str.ReadLine(fin1); while (1) { if (!fin0 || !fin1) break; double Emc, Size, Zd, Dist, Disp, Slope, M3L, Leakage, Width, Length, Eest; fin0 >> Emc >> Size >> Zd >> Dist >> Disp >> Slope >> M3L >> Leakage >> Width >> Length; fin1 >> Eest; h2svse.Fill(Emc, Size); h2est.Fill( Emc, Eest); h2bias.Fill(Eest, Eest-Emc); h2lin.Fill( Eest, (pow(10, Eest)-pow(10, Emc))/pow(10, Emc)); h2size.Fill( Size, Eest-Emc); h2dist.Fill( Dist, Eest-Emc); h2slope.Fill(Slope, Eest-Emc); h2zd.Fill( Zd, Eest-Emc); } TF1 fx("f", "x", -100, 100); TF1 f0("f", "0", -100, 100); TCanvas *canv = new TCanvas("Canvas", "Energy estimation", 800, 900); canv->Divide(2,4); canv->cd(1); gPad->SetTopMargin(0.005); gPad->SetGridy(); h2size.SetStats(kFALSE); h2size.SetXTitle("lg(E_{mc}/GeV)"); h2size.SetYTitle("lg(E_{est}/GeV)-lg(E_{mc}/GeV)"); h2size.DrawCopy("colz"); f0.DrawCopy("same"); canv->cd(3); gPad->SetTopMargin(0.005); gPad->SetGridy(); h2dist.SetStats(kFALSE); h2dist.SetXTitle("Dist / degree"); h2dist.SetYTitle("lg(E_{est}/GeV)-lg(E_{mc}/GeV)"); h2dist.DrawCopy("colz"); f0.DrawCopy("same"); canv->cd(5); gPad->SetTopMargin(0.005); gPad->SetGridy(); h2slope.SetStats(kFALSE); h2slope.SetXTitle("Slope / ns/degree"); h2slope.SetYTitle("lg(E_{est}/GeV)-lg(E_{mc}/GeV)"); h2slope.DrawCopy("colz"); f0.DrawCopy("same"); canv->cd(7); gPad->SetTopMargin(0.005); gPad->SetGridy(); h2zd.SetStats(kFALSE); h2zd.SetXTitle("Zenith Distance / degree"); h2zd.SetYTitle("lg(E_{est}/GeV)-lg(E_{mc}/GeV)"); h2zd.DrawCopy("colz"); f0.DrawCopy("same"); canv->cd(2); gPad->SetTopMargin(0.005); gPad->SetGridy(); h2svse.SetStats(kFALSE); h2svse.SetXTitle("lg(Size)"); h2svse.SetYTitle("lg(E_{mc}/GeV)"); h2svse.DrawCopy("colz"); fx.DrawCopy("same"); canv->cd(4); gPad->SetTopMargin(0.005); gPad->SetGridy(); h2est.SetStats(kFALSE); h2est.SetXTitle("lg(E_{mc})"); h2est.SetYTitle("lg(E_{est}/GeV)"); h2est.DrawCopy("colz"); fx.DrawCopy("same"); canv->cd(6); gPad->SetTopMargin(0.005); gPad->SetGridy(); h2bias.SetStats(kFALSE); h2bias.SetXTitle("lg(E_{est}/GeV)"); h2bias.SetYTitle("lg(E_{est}/GeV)-lg(E_{mc}/GeV)"); h2bias.DrawCopy("colz"); f0.DrawCopy("same"); canv->cd(8); gPad->SetTopMargin(0.005); gPad->SetGridy(); TGraph grlin; TGraph grlog; for (int x=1; x<h2bias.GetNbinsX(); x++) { TH1D *p = h2bias.ProjectionY("_py", x, x); if (p->GetRMS()>0) grlog.SetPoint(grlog.GetN(), h2bias.GetXaxis()->GetBinCenter(x), pow(10, p->GetRMS())-1); delete p; p = h2lin.ProjectionY("_py", x, x); if (p->GetRMS()>0) grlin.SetPoint(grlin.GetN(), h2lin.GetXaxis()->GetBinCenter(x), p->GetRMS()); delete p; } grlog.SetMarkerColor(kBlue); grlin.SetMinimum(0); grlin.GetHistogram()->SetXTitle("log(E_{est}/GeV)"); grlin.GetHistogram()->SetYTitle("RMS"); grlin.DrawClone("A*"); grlog.DrawClone("*"); }
Result
And here are the plots which were produced:
Attachments (1)
- Result.png (37.7 KB ) - added by 8 years ago.
Download all attachments as: .zip